rhapsodi, R haploid sperm/oocyte data imputation, is an R package meant to work with very low-coverage single-cell DNA sequencing data of gametes originating from a single diploid donor. Specifically, rhapsodi (1) phases the diploid donor haplotypes, (2) imputes missing gamete genotypes, and (3) discovers gamete-specific meiotic recombination events.
rhapsodi can be installed with devtools, as follows:
library(devtools)
install_github("mccoy-lab/rhapsodi")
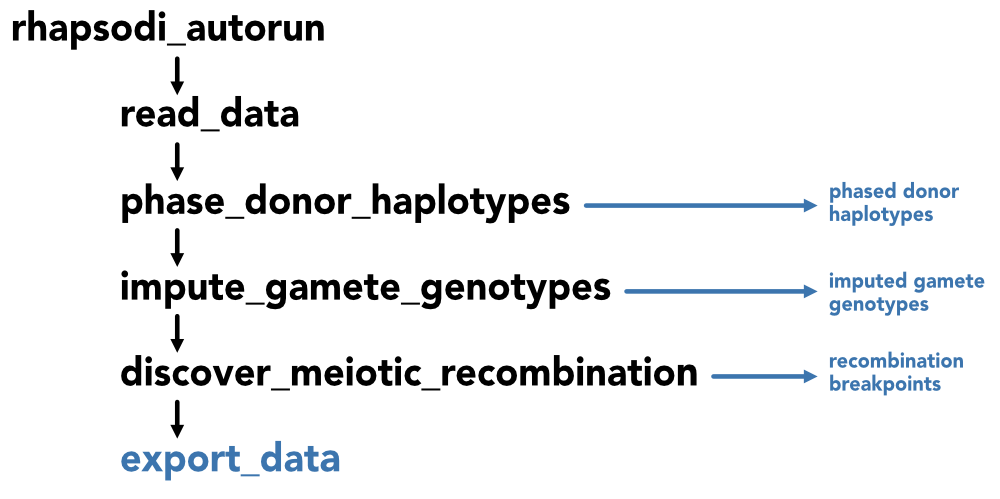
The full rhapsodi analysis pipeline can be driven by the rhapsodi_autorun function that imports the data (read_data), runs the 3 main stages of analysis (phase_donor_haplotypes, impute_gamete_genotypes, & discover_meiotic_recombination), and finally exports the data from each stage of analysis in a named list (export_data). Alternatively, each of these steps can be run by themselves, however, the 3 stages must be run in order as the output of each is used in the next step.
The rhapsodi vignette can be viewed here: rhapsodi vignette